Apologies, but no results were found.
Paul Shipley
Associate Professor
Biochemistry and Molecular Biology, Chemistry
Other Titles: Associate Dean, College of Graduate StudiesOffice: OM2 113
Phone: 250.807.8749
Email: paul.shipley@ubc.ca
Graduate student supervisor

Research Summary
Organic chemistry; natural products biosynthesis; the biochemistry of secondary metabolism; nuclear magnetic resonance spectrometry; rational modification of biosynthetic pathways in bacteria to produce novel compounds with the potential to be pharmacologically relevant.
Courses & Teaching
Organic chemistry; structural spectrometry; bioorganic chemistry
Degrees
PhD, University of Washington
Research Interests & Projects
Our lab focuses on describing the chemistry of medicinal plants and bacteria to investigate the chemical differences between species and samples. This is used to discover biological activities, optimize natural health product formulation, identify adulterated products, and classify species by their chemistry.
Metabolomic Analysis of Medicinal Plants by NMR
Nuclear magnetic resonance (NMR) metabolomics is an emerging field where NMR spectra are used to provide a picture of the total chemistry of a sample, as opposed to its traditional use with purified compounds. My lab has developed new approaches to defining the complex metabolome of plant samples. It has recently been estimated that an average plant tissue contains up to 30,000 distinct phytochemicals but chemical methods to define this diversity are lacking.
My students are investigating NMR-based chemical approaches to medicinal plant analysis for a variety of different species. We are developing methods to compare different species, extraction methods, and growing conditions with a combination of one and two dimensional NMR spectrometry and statistical analysis of the data sets. We are comparing these results with both LC/MS metabolomic analysis and chromatographic separation and quantification of known metabolites.
Recent Projects
Hawthorn (Crataegus spp.) have well documented cardioprotective properties. We have looked at the chemical profiles of North American, European, and Asian hawthorn species from a chemotaxonomic perspective, where we are able to distinguish hawthorn species from different clades, and to identify which compounds give rise to that variance, some of which were compounds that have had demonstrated cardioprotective activities.
Cranberry spp. has a demonstrated efficacy against urinary tract infections. Using NMR metabolomic analysis we have profiled different extraction conditions. Initial investigations showed that the profiles between landraces and growing conditions were not significantly distinguished, leading to the model improvement work described below.
Advanced Statistical & Methodological Tools for Model Improvement
In our investigations of Crataegus spp. chemistry we have investigated optimization of the type of NMR experiment that gives the best discrimination between sample types, finding that while skyline projection of 2D JRES data simplifies the spectrum considerably, we are able to build far more robust statistical models using standard 1D proton data sets. This is true of both more traditional models and with machine learning algorithms. We have been able to generate discriminating models that clearly distinguish between North American and European species, and cladistics separations are clear as well.
LC/MS based metabolomic analyses often suffer from statistical significance being incorrectly identified in regions of baseline noise. We have developed several statistical tools for discriminating between true and false positives in significance analysis. As a model system, we compared Vaccinium species to each other, as well as a comparison of commercial varietals. With samples with such a high degree of chemical similarity, we were able to use univariate statistical analysis to refine our data sets prior to multivariate analysis. This allowed us to describe how chemistry varies in a significant way between varietals grown in close to identical growing conditions. We are currently working to apply these techniques to NMR data sets as well.
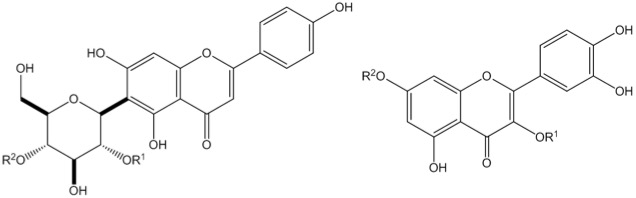
Diagram 1: Plant phenolic compounds found in Hawthorn that have cardioprotective activities.
Selected Publications & Presentations